Bat Detective - Deep Learning Tools for
Bat Acoustic Signal Detection
Oisin Mac Aodha, Rory Gibb, Kate E. Barlow, Ella Browning, Michael Firman, Robin Freeman, Briana Harder, Libby Kinsey, Gary R. Mead, Stuart E. Newson, Ivan Pandourski, Stuart Parsons, Jon Russ, Abigel Szodoray-Paradi, Farkas Szodoray-Paradi, Elena Tilova, Mark Girolami, Gabriel J. Brostow, and Kate E. Jones
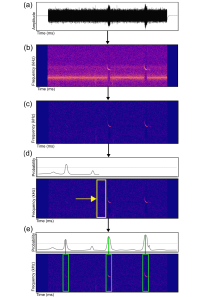
Passive acoustic sensing has emerged as a powerful tool for quantifying anthropogenic impacts on biodiversity, especially for echolocating bat species. To better assess bat population trends there is a critical need for accurate, reliable, and open source tools that allow the detection and classification of bat calls in large collections of audio recordings. The majority of existing tools are commercial or have focused on the species classification task, neglecting the important problem of first localizing echolocation calls in audio which is particularly problematic in noisy recordings.
We developed a convolutional neural network based open-source pipeline for detecting ultrasonic, full-spectrum, search-phase calls produced by echolocating bats. Our deep learning algorithms were trained on full-spectrum ultrasonic audio collected along road-transects across Europe and labelled by citizen scientists from www.batdetective.org. When compared to other existing algorithms and commercial systems, we show significantly higher detection performance of search-phase echolocation calls with our test sets. As an example application, we ran our detection pipeline on bat monitoring data collected over five years from Jersey (UK), and compared results to a widely-used commercial system.
Our detection pipeline can be used for the automatic detection and monitoring of bat populations, and further facilitates their use as indicator species on a large scale. Our proposed pipeline makes only a small number of bat specific design decisions, and with appropriate training data it could be applied to detecting other species in audio. A crucial novelty of our work is showing that with careful, non-trivial, design and implementation considerations, state-of-the-art deep learning methods can be used for accurate and efficient monitoring in audio.
BibTeX
@inproceedings{batdetect18,
title = {Bat Detective - Deep Learning Tools for Bat Acoustic Signal Detection},
author = {Mac Aodha, Oisin and Gibb, Rory and Barlow, Kate and Browning, Ella and
Firman, Michael and Freeman, Robin and Harder, Briana and Kinsey, Libby and
Mead, Gary and Newson, Stuart and Pandourski, Ivan and Parsons, Stuart and
Russ, Jon and Szodoray-Paradi, Abigel and Szodoray-Paradi, Farkas and
Tilova, Elena and Girolami, Mark and Brostow, Gabriel and E. Jones, Kate.},
journal={PLOS Computational Biology},
year={2018}
}
Acknowledgements
We are enormously grateful for the efforts and enthusiasm of the amazing iBats and Bat Detective volunteers, for the many hours spent providing valuable annotations. We would also like to thank Ian Agranat and Joe Szewczak for useful discussions and access to their systems. This work was supported financially through the Darwin Initiative (Awards 15003, 161333, EIDPR075), Zooniverse, the People’s trust for Endangered Species, Mammals Trust UK the Leverhulme Trust (Philip Leverhulme Prize for KEJ), NERC (NE/P016677/1), and EPSRC (EP/K015664/1 and EP/K503745/1).

 paper
paper
 code
code
 data
data